-Search query
-Search result
Showing 1 - 50 of 134 items for (author: fry & ee)
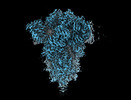
EMDB-18807: 
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
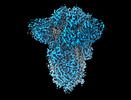
EMDB-18808: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1c: 
SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1d: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-16680: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

PDB-8cin: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-16676: 
Delta-RBD complex with BA.2-07 fab, SARS1-34 fab and C1 nanobody
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-43049: 
Cryo-electron tomogram of mitochondria from cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43050: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43051: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43052: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43053: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43054: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43055: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43056: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43057: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43058: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43059: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43060: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43061: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Luce BE, Lugo CM, Chao LH

EMDB-43062: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion
Method: electron tomography / : Fry MY, Navarro PP, Ananda VY, Ge Y, McDonald JL, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43063: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43064: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH
EMDB-43065: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43066: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43067: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43068: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43069: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-43070: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1
Method: electron tomography / : Navarro PP, Fry MY, Ananda VY, Ge Y, McDonald JL, Ali I, Hakim P, Lugo CM, Luce BE, Chao LH

EMDB-16678: 
BA.2-07 FAB IN COMPLEX WITH SARS-COV-2 BA.2.12.1 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-15971: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8bcz: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-26809: 
KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate
Method: single particle / : Spangler CJ, Skrajna A, Foley CA, Budziszewski GR, Azzam DN, James LI, Frye SV, McGinty RK

EMDB-26810: 
KDM2B-nucleosome complex stabilized by H3K36C-UNC8015 covalent conjugate
Method: single particle / : Spangler CJ, Skrajna A, Foley CA, Budziszewski GR, Azzam DN, James LI, Frye SV, McGinty RK

PDB-7uv9: 
KDM2A-nucleosome structure stabilized by H3K36C-UNC8015 covalent conjugate
Method: single particle / : Spangler CJ, Skrajna A, Foley CA, Budziszewski GR, Azzam DN, James LI, Frye SV, McGinty RK

EMDB-15920: 
Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55)
Method: single particle / : Stuart DI, Ren J, Zhou D, Qin L

EMDB-15930: 
Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion
Method: single particle / : Stuart DI, Ren J, Qin L, Zhou D

EMDB-15725: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3
Method: single particle / : Bahar MW, Fry EE, Stuart DI
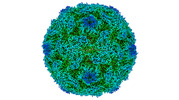
EMDB-15726: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril
Method: single particle / : Bahar MW, Fry EE, Stuart DI
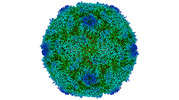
EMDB-15727: 
Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8ayx: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8ayy: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8ayz: 
Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17
Method: single particle / : Bahar MW, Fry EE, Stuart DI
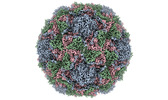
EMDB-15543: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8).
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8anw: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8).
Method: single particle / : Bahar MW, Fry EE, Stuart DI

EMDB-15234: 
Structure of self-assembling engineered protein nanocage (EPN) fused with hepatitis A pX protein
Method: single particle / : Duyvesteyn HME, Stuart DI
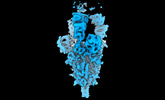
EMDB-14885: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14886: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14887: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
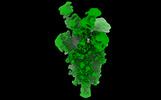
EMDB-14910: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
